Introduction
Welcome to the ultimate guide to how cloud computing enables seamless research experience. In this blog, we’re going to look at how cloud computing has changed the way that researchers collaborate. We’re going to look at some examples as to how to improve that collaboration. And we’re going to look at some of the current sticking points and hindrances when it comes to users collaborating. And then we’re going to try and find ways to improve and enhance that collaboration so that researchers find it easier to work amongst each other when it comes to distributed computing.
Researchers have often worked in very siloed environments, environments that make it incredibly difficult to share and collaborate in anything close to real time. Working remotely often makes it prohibitive to share data in any way other than Jupyter notebooks or Excel spreadsheets. In reality, I’m doing them a disservice, but quite often they are the ways that people, researchers share their data or they sit on their data until it’s ready for publication and then disseminate it at that point in a published paper, which is entirely fine. But there’s a number of cases where we can really improve upon the way that research is done, especially when researchers may be sat the other side of the globe to make their lives easier, more effective, reduce costs and overhead, and really foster that sort of real time understanding of data.
Modern technology, open source and open standards allow for data to be shared in highly optimized ways, globally using tooling available on all large and most of the not so large cloud providers. Object storage is cheap, reliable storage and these days drives the storage layer of so many data processing solutions. Alongside the cheap storage, compute either serverless or ondemand means you only pay for what you use, when you use it. Combining these two very powerful features, means that researchers and work on a budget but still collaborate with others around the globe.
Understanding Cloud Research Fundamentals
So, a little history lesson, if we roll back to the 1960s, the advent of computing in the mainstream, and also specifically the internet and cross-border collaboration, this all started in 1969 with ARPANET, which was the precursor to the internet. We roll on through the 70s and 80s, computers improved, the way that networks worked improved, and then the World Wide Web, as we know it, effectively launched in 1991, connecting millions of machines across the world. This then snowballed into distributed computing, cloud computing, and the large cloud service providers that we know today with Amazon Web Services, Microsoft Azure, Google Cloud, et cetera, et cetera. There’s a number of players offering almost unlimited compute in a cloud environment.
This has, of course, then led to an explosion of distributed researchers and researchers working around the globe on shared problems that fundamentally can change the world that we live in. They can change policies. They can change medical understanding. There’s global research that is done on a daily basis with teams around the globe, but the way that researchers often work is still relatively antiquated and hasn’t necessarily changed a great deal since they started doing their research. For data scientists and researchers alike, notebook-type environments that give you access to a SQL interface or a Spark server or something of that nature, or even just a Pandas environment where you can process tabular data in a scriptable manner, is the bread and butter of the way that people do research. If they can’t do that, then often Microsoft Excel is a tool that is leveraged on a regular basis to allow researchers to process their data. The problem with this, though, is that sharing this data in real time and also sharing knowledge, understanding, and expectations around this data is not necessarily something that is collaborative when you want to work with teams that are distributed around the globe. These days, universities and research institutes will pick different universities distributed around the globe because it gives them access to different grants and funds and potential legal avenues that allow them to do that research. But they need a way to be able to visualize and conceptualize the data that they’re processing so that they can collaborate effectively, reducing cost and improving the time it takes them to do that research. And so, we want to look at ways and means for us to be able to improve that process and really allow researchers access to important data points in a way that allows them to share what they’re seeing and ask the right questions of researchers to help validate what they’re trying to achieve.
The majority of cloud interactions are web based, either via a UI or a CLI of some sort triggering. a process on the cloud that then achieves something. The beauty of modern web frameworks is that you can visualize almost any dataset, all you need is the clarity of thought to know what you want to see and how you want to interact with it.

Getting Started with Cloud based Research Collaboration
So where does cloud based research collaboration really begin? What services are out there? What do we need to hand crank? How do we get different researchers involved?
Firstly I would like to look at a few solutions, and by no means all of them, that we can spin up, pretty much at 1 click of a button. Hosted in the cloud and designed for researchers, data scientists and engineers to run their processes over their code.
Pre Canned Cloud based solutions
Databricks
Lets start with what is, if you look at any online channel at the moment, the main player in the online data processing sector. Databricks has been around for a long time but recently has really driven massive growth in this space alongside the advent of LLMs, running models in the cloud, and other governance features, Databricks is a managed Apache Spark platform which provides users with the ability to load up data and then process it using either PySpark, Scala or SQL. The main interface for doing so is a notebook type interface, and this allows for scripted access to run data transformation and visualizations over your data. There is also a dashboard solution and data vizualization tool which allows for more end user driven access to the data that has been manipulated elsewhere in the platform.
Snowflake
Next on the list is Snowflake, which as of late 2024 seems to be Databricks most close competitor. Snowflake is a cloud-based data platform that enables seamless storage, processing, and analysis of large volumes of data. Unlike traditional databases, Snowflake is designed to handle vast amounts of data from diverse sources, which makes it ideal for distributed research. With its architecture, multiple users can access and query data simultaneously without performance issues.
Researchers from different locations can collaborate in real-time, making data-driven decisions quickly and efficiently. Snowflake’s powerful data-sharing capabilities allow for secure and controlled data access, ensuring that sensitive information remains protected. It also integrates easily with various data analysis and visualization tools, streamlining the research process.
Snowflake simplifies big data management and collaboration, making it easier for distributed research teams to work together, draw insights, and advance their projects with speed and security.
Azure Lakehouse
An Azure Lakehouse is an innovative data architecture that unifies data lakes and data warehouses to provide a seamless integration of structured and unstructured data. Traditionally, data lakes store vast amounts of raw data, while data warehouses store processed, structured data optimized for analytics. Azure Lakehouse bridges these two by providing a single platform where diverse data types can coexist and be queried with high efficiency.
For collaborative research, an Azure Lakehouse offers significant advantages. Researchers can ingest and store massive datasets from various sources without worrying about preprocessing data into a specific format. The integrated environment allows for easier data sharing and joint analysis, fostering collaboration among multidisciplinary teams. Furthermore, the lakehouse supports powerful analytics and machine learning workloads, enabling researchers to derive deeper insights and accelerate discoveries.
With its ability to streamline data management and provide a unified analytical framework, an Azure Lakehouse can greatly enhance the productivity and coordination of collaborative research projects.
Sagemaker
Next on the list is SageMaker. AWS SageMaker is a powerful cloud service that makes machine learning accessible and easy, even for beginners. It can significantly boost collaborative research in several ways. Firstly, SageMaker provides a shared online workspace where multiple researchers can work together from anywhere in the world. This makes it easy to share data, models, and insights in real-time, enhancing teamwork.
Secondly, SageMaker simplifies complex machine learning tasks by offering pre-built algorithms and user-friendly tools, like notebooks. You don’t need to be a machine learning expert to use it; even novices can build and train models with ease. SageMaker also handles all the behind-the-scenes technical stuff, like managing servers and data storage. This way, researchers can spend more time focusing on their research rather than worrying about technical details.
Google Colab
Google Colab, short for Google Collaboratory, is a cloud-based platform that provides a free environment for coding and data analysis. It is particularly popular among researchers and data scientists for its convenience and collaborative features. Essentially, Google Colab allows users to write and execute Python code in a Jupyter Notebook interface, directly through a web browser, eliminating the need for complex local setups.
One of the key advantages of Google Colab is its support for real-time collaboration on notebooks. Multiple users can simultaneously work on the same document, making it easy to share code, data, and results. This feature fosters seamless teamwork, allowing researchers from different locations to contribute to a project with ease.
Additionally, Google Colab provides access to powerful computational resources, including GPUs and TPUs, at no cost. This capability is particularly beneficial for processing large datasets and training machine learning models. By leveraging these resources, researchers can expedite their data processing tasks without investing in expensive hardware.
Roll Your Own
There are a whole host of open source tools out there to help you with your data maniulation and sharing but I’m going to run through 3 today. They are Jupyter Labs, notebooks, Apache Superset, dashboards and DuckDB, data storage. There are an awful lot more solutions out there, I have just chosen one from each area so we don’t have another huge list!
Jupyter Labs
Similar to Google Collab, JupyterLab is an advanced, open-source web-based interface for Jupyter notebooks, making it an invaluable tool for researchers and data scientists. Jupyter notebooks themselves allow users to create and share documents containing live code, equations, visualizations, and narrative text. JupyterLab enhances this by providing a more flexible and integrated environment, supporting multiple file types and workflows.
For remote collaboration, JupyterLab shines with its ability to work seamlessly over the web. Researchers can access, edit, and share their notebooks from any location with an internet connection, facilitating real-time collaboration. By using platforms like GitHub or cloud services like Google Colab, teams can co-develop and analyze data sets, run simulations, and visualize results together, irrespective of geographical barriers. Its support for numerous programming languages further broadens its appeal. JupyterLab’s robust ecosystem significantly improves efficiency and cooperation on complex data science projects.
Apache Superset
Apache Superset is an open-source data exploration and visualization platform that can significantly enhance collaboration among remote researchers. By providing a unified interface, Superset enables users to access, visualize, and share complex datasets seamlessly, eliminating geographical barriers. Researchers can create interactive dashboards and charts, making it easy to interpret data patterns and trends. These visualizations can be easily shared with team members or publicly, fostering a more collaborative environment.
Moreover, Superset supports a variety of data sources, including SQL databases and big data engines, allowing researchers to work within a unified framework without the need for extensive data migrations. The platform’s SQL editor and intuitive drag-and-drop interface make it accessible to users with varying levels of technical expertise, thus promoting inclusivity. Real-time collaboration features, such as shared dashboards and comment functionalities, ensure that team members remain synchronized, facilitating more effective and timely decision-making. Overall, Apache Superset empowers remote research teams to work more cohesively and efficiently.
DuckDB
DuckDB is a high-performance, in-process SQL database management system designed for data analysis. It can significantly enhance collaboration among researchers working in a cloud environment. Unlike traditional database systems, DuckDB is lightweight and easy to integrate within any application, making it ideal for handling complex data queries efficiently.
In a cloud setting, researchers can utilize DuckDB to store, query, and analyze massive datasets without the need for dedicated database servers. DuckDB’s in-process architecture allows for seamless data processing directly within the researcher’s preferred programming environment, such as Python or R. This facilitates real-time data analysis and reduces latency.
Furthermore, DuckDB supports efficient data sharing and version control, making it easier for teams to collaborate. Researchers can share scripts and queries, ensuring consistent and reproducible results. Its compatibility with various data formats also means that researchers can combine diverse datasets from different sources effortlessly, fostering a collaborative and integrative research approach.
How do you choose?
This is a very good question and it depends on a number of things. The 2 biggest drivers for the solution are budget and data volume.
On the budget front, you have to think about it in a few different ways. Firstly, by having your operations hosted in the cloud you’re removing the need for a lot of the engineering and support you’d otherwise need if it were hosted elsewhere, but of course in hosting this platform somewhere you’re costing more than it would do otherwise if the data was just on a laptop. The fact that we’re here discussing collaboration and the use of cloud services assumes that this has at least crossed your mind. The other cost terms often though are determined by, frequency of access of the data, or data processing volume. The size of the data you have in your warehouse. And lastly the tooling you chose to use. For example, if you’re an Azure user, Databricks is a lot more expensive than Azure Lakehouse, but equally it has a lot more features.
These two though are pretty complex to get started with and would likely require some support from an Cloud Engineer or similar to get the research team going. So what else is out there? Google Colab is a cost effective and relatively simple hosted option to get going with. AWS Sagemager is reasonably heavyweight but if you already have an AWS account it might be a reasonably easy way to get access to hosted notebooks.
The benefit of most of these services is that you pay for the compute you use. They all use Object Storage (S3-esque) on the backend and so you pay reasonably low costs for the data at rest. You pay for the compute. Depending on the size of the cluster, the length of time its running and the amount of data you’re sticking through it will have a large determination to the final monthly cost. As your data volume increases, depending on how the data is processed your compute costs may also increase, or, they may remain pretty constant throughout a month and you’re just paying for more finished storage.
When evaluating costs also pay attention to what you plan to do with the storage. Most table formats these day are backed by the Parquet table structure which compresses a lot of textual information, keeping the storage size down. Databricks though have championed the “medallion architecture”and this involves various copies of data in bronze and silver layers with differing amounts of manipulation done on them. Of course its absolutely not mandatory to use this architecture but if you do, it needs factoring into the costs.
Advanced Strategies for using the Cloud for Research
Of course it doesn’t need to end there. Another aspect of research sharing is, sometimes its appropriate to break the mould and really go bespoke. So I will tell you how we built Pixlise and how sometimes offering up a completely bespoke alternative is the way to go.
Mars Rovers and everything in between
We’ve worked with NASA JPL for a number of years and in that time we’ve built a lot of research platforms. Most of them aren’t fully featured, their proof of concepts to test out ideas and find out how best to complete a specific task before the project owner goes and applies for more funding. But occasionally a fully funded project would fall into your lap. And in this case it was a tool called Pixlise for the NASA Perseverance Mars Rover.
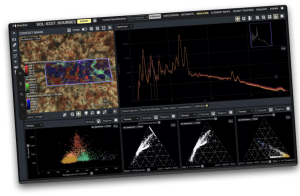
Pixlise is an open source tool thats designed to process and visualize X-Ray spectrometry data. I don’t pretend to know much about X-Ray spectrometry but it doesn’t mean you can’t build a tool around it. So in this case we were tasked with building a tool that helped a distributed group of scientists find out what the chemical makeup of rocks on Mars was. All normal run of the mill stuff!
Our team was blessed to have a manager who had a PhD in human computer interaction, and a UX researcher who was very good. And so they really dug into the pain points of the researchers, working out what was important to them. Of course at the start of the project there was a general reluctance to shift away from their existing Excel workflow, but part of the problem there was that it usually took about 30 hours and 1 persons laptop to do the processing work from a dataset, and the team were being asked for almost realtime answers on whether they would stay or go at a certain site depending on the scan output, so 30 hours was clearly unacceptable.
So first we had to figure out what we were going to build, what the architecture would look like and how we could drive this innovation with what was effectively a 2 developer team. Performance was obviously one aspect, another factor was how us as a development team would build this application with JPL in LA, myself in London and the other developer in Melbourne. It was going to be a logistical challenge, so we developed a platform that kept it simple, allowed us to iterate and work on building this application out.
Iteration was key because core to this whole project was researcher acceptance and their belief that this application stack would be a large improvement on their existing workflows. If they didn’t believe that then we’d not build the tool they desired and they would continue to do work in silos.
The outcome from the project was a tool that took the data from the Mars Rover and got the processing down to 20 minutes, usually a lot less, which gave the researchers what they needed. But it also turned into a large collaboration platform, they shared, internally, and then externally, data sets, views, analysis, formula, findings, you name it, on the platform, in the cloud. Reducing that initial instinct to dump a CSV to the laptop and do things in Excel. We even implemented a custom scripting language that allowed them to write additional formula and similar for data queries that didn’t have an answer to what they needed already at hand.
Asking the important questions
The key to the success of the Pixlise project was that the UX research was really top notch, and so many questions were asked to the researchers about what they needed and dig deeply, ensure that what they were telling you was the finished product and not some middle ground they felt comfortable asking for before exporting the data and finishing it off locally later.
Often end users, regardless of their background will tell you somewhere approaching what they want but quite often a middle ground where they may think its a bit of a hassle to ask for it all so something that gets you 2/3 of the way there to then finish off the rest on their own at a later date. Its important to really dig into the use cases and how they will end up interacting with any new components that are designed.
The complementary piece of asking the important questions is also listening to the feedback and ensuring you don’t waste your time or the interviewees time but taking the valid feedback and working it into your product roadmap. In doing this you will ensure you keep the end users engaged and interested in what you have to offer.
Research citations and data access
Pixlise has now been cited in a number of papers, in part due to the flexibility of the tooling we built and the researchers enjoyment in using a tool that really sped up the time it took to get the answers they needed.
Ground breaking research about rock composition on the surface of Mars and the makeup of those rocks has been carried out and the data it then made accessible to other researchers, within the tooling so they can see and validate the research that has been done.
Whats more, the software was then released open source, and so researchers can validate the computation, the data, the processing and really understand how the whole platform works end to end.
The Future of Cloud based Research
Cloud based data processing
Cloud based data processing isn’t going anywhere. There is so much scope in the tooling, the scale and the use cases that Cloud computing in general is just how most of us operate these days.
Don’t forget though, and this is very important, just because you have cloud scale compute at your fingertips doesn’t mean you need to architect for cloud scale data processing environments. You likely don’t have a Youtube sized dataset, in which case, adjust accordingly.
Sometimes its as easy to spin up an EC2 server, ensure it can connect to an S3 bucket and stick your data in there. Sometimes though it doesn’t hurt to use something like Databricks, you only pay for the compute you use after all, and so getting that environment configured and accessible is usually a one time thing, after that you and your fellow researchers can collaborate of shared datasets for the length of the project.
There’s also no real right answer, a lot of this comes down to decisions made at organisational level. Do we already use a cloud provider? If so, what do they offer? Can we use it? The other important note is, experiment. Experiment with different tools, most cloud providers will offer a useable free tier, and you can really dig into the offering and how it might work with you and your team.
Custom UI and interactions
Not every project needs a custom UI, I’d go so far as saying most projects don’t need a custom UI, but some, when time and budget allows should consider investing in one. The project I’m currently working on is a hybrid type of environment, we’ve got some custom dashboards, we’ve got some hosted notebooks and we’ve got scalable cloud compute underpinning it all.
There is a middle ground though with platforms like Superset, Quicksight or PowerBI if you want to got the commercial route, that offer a degree of flexibility in terms of graphics on screen whist still being a supported application that you can manage.
Taking that time, and talking to researchers about what they want to really see is what can really make or break the usability of a project and its worth investing the energy up front and then iterating over it to really fine tune what you’re offering.
AI?!!
I wouldn’t be able to write an entire article without at least discussing AI briefly. AI covers a lot of topics, LLMs which are all the rage today are one thing, but so is more traditional Machine Learning modelling. The cloud and its reasonably infinite resource will forever be the home of model training and deployment for a lot of us mortals as we don’t have a GPU cluster sat under our desk. Over time the models we will use will evolve and get more complex, the use of LLMs and so on will increase as we build more advanced functionality into the backend of our research applications.
Frequently Asked Questions
- What are the main benefits of cloud computing for scientific research?
- Cloud computing offers several key advantages for scientific research:
- Increased computational power: Cloud platforms provide access to high-performance computing (HPC) resources that can handle complex simulations, large-scale data analysis, and demanding computational tasks.
- Cost-effectiveness: Cloud services operate on a pay-as-you-go basis, allowing researchers to avoid large upfront investments in hardware and infrastructure.
- Scalability: Researchers can easily scale computing resources up or down based on project needs.
- Collaboration: Cloud platforms facilitate data sharing and real-time collaboration among researchers across different locations.
- Accessibility: Cloud resources can be accessed from anywhere with an internet connection, enabling remote work and increasing flexibility
- Cloud computing offers several key advantages for scientific research:
- What types of cloud services are most useful for scientific research?
- Several cloud services are particularly valuable for scientific research:
- High-performance computing (HPC) resources: For running complex simulations and data analysis.
- Data storage and management: For securely storing and organizing large datasets.
- Virtual machines: For running specialized software and creating custom research environments.
- How can cloud computing improve the quality and quantity of research?
- Cloud computing can enhance research in several ways:
- Enabling more advanced analyses: Access to powerful computing resources allows researchers to tackle more complex problems and derive more accurate results.
- Increasing research output: Cloud resources can speed up computations and allow for parallel processing, enabling researchers to complete more work in less time.
- Facilitating reproducibility: Cloud platforms can provide consistent environments for running analyses, making it easier to reproduce and validate research results
- Web hosting: For sharing research tools and results with the scientific community
- Cloud computing can enhance research in several ways:
- What are the potential risks or challenges of using cloud computing for research?
- While cloud computing offers many benefits, researchers should be aware of potential challenges:
- Data security and privacy: Storing sensitive research data in the cloud may raise concerns about data protection and compliance with regulations.
- Cost management: While cloud services can be cost-effective, researchers need to carefully monitor usage to avoid unexpected expenses.
- Learning curve: Adopting cloud technologies may require new skills and training for research teams.
- Data transfer and network limitations: Moving large datasets to and from the cloud can be time-consuming and may be limited by network bandwidth
- While cloud computing offers many benefits, researchers should be aware of potential challenges:
- How can researchers get started with cloud computing?
- To begin using cloud computing for research:
- Assess your computational needs and identify which aspects of your research could benefit from cloud resources.
- Explore cloud platforms that cater to scientific research, such as those offered by major providers or specialized academic cloud services.
- Start with small-scale projects to familiarize yourself with cloud technologies and best practices.
- Consider seeking training or consulting with cloud computing experts to optimize your use of cloud resources
- To begin using cloud computing for research:
- Are there any special considerations for researchers using cloud computing?
- Researchers should keep in mind:
- Data sovereignty: Be aware of where your data is stored, especially for sensitive or regulated research data
- Export control laws: Ensure compliance with regulations that may restrict where certain types of research data can be stored or processed
- Funding and budgeting: Consider how to incorporate cloud computing costs into research grant proposals and budgets
- Long-term data management: Plan for data storage and accessibility beyond the duration of individual research projects
- Researchers should keep in mind:
By understanding these aspects of cloud computing in scientific research, researchers can make informed decisions about how to leverage cloud resources to enhance their work and drive innovation in their fields.
Conclusion
There is a lot of complexity in the cloud and it can certainly make life trickier in the short run, removing the “this is how we’ve always done it aspect”. But with that complexity comes great power, both for the user but also in terms of almost unlimited compute and storage for most peoples needs. The advance in cloud based technologies means that most things you do locally already exist in some hosted service on the cloud and if they don’t you can spin up a virtual machine or virtual desktop and do what you do locally on larger machines in a cloud environment. But thats not all, and not the point of this article, what it also provides is the ability for researchers to be able to share work, collaborate and get access to different datasets, more quickly and easily.
Depending on budget there are obviously a variety of different ways to go. Smaller projects may chose to just leverage object storage, low cost VMs or containers and cloud based ETL services. Large projects might go the full custom application design route that can really up the researchers game, showing them what they really need to see in almost real time.
The options really are endless, the limit is what you want and the time and effort it would take to implement it. But also consider this. Is this a one shot piece of research? Are you going to come back and do it again, if so how can you leverage cloud environments to really allow for repetition, at scale. This will make those repeated data extracts and analysis much more cost effective, driving down the budget and costs for future pieces of research. If you’ve written code, can you open source it? Can you make that code available to other researchers? Allowing them a head start on the their own work, delivering greater value for money for projects in the future.
If you feel like any of this blog has been useful there are a number of other resources out there but do check out our work on Pixlise. What we’re doing at the Accelerator and what others are doing in this field. It really is eye opening. And as ever if you feel like this resonates and you want help implementing something similar, regardless of size, feel free to get in touch!
If your company could use our expert knowledge in deploying and scaling systems, then book an introductory call and find out how Spicule can help.
Thanks for reading Idea Ignition: Fueling Startups from Concept to Cloud! Subscribe for free to receive new posts and support my work.
